Abstracted Protein Simulator
The Abstracted Protein Simulator models proteins interacting in 3D, at a mesoscopic level. It was designed for studying the self-assembly and operation of protein machines containing 10s or 100s of proteins, and operates at a coarser temporal and spatial scale than most simulators.
Our main model system is the development of the post synaptic density, a complex assembly of ion channels and cytoskeleton which is a critical part of learning and memory in the synapse. We have also been testing the simulator on self assembly of simple viruses from their constituent proteins, since more complete molecular data sets are available for viruses than neurons.
- The model specification is an XML file which includes details of protein shapes, binding sites, interaction and state change probabilities. The simulation algorithm is based on monte-carlo techniques.
- The simulator provides a graphical user interface for entering data on protein structure and bonding.
- The simulation is shown running in a 3D animation.
Example
Protein structures can be input in .pdb format from databases.

|
|
The structure can be simplified by grouping by amino acid...

|
|
...or clustered into a simpler structure.

|
|
Bonds can be defined and added to the protein template.
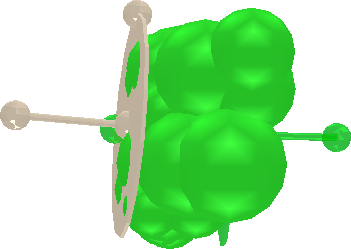
|
|
The simulation is run
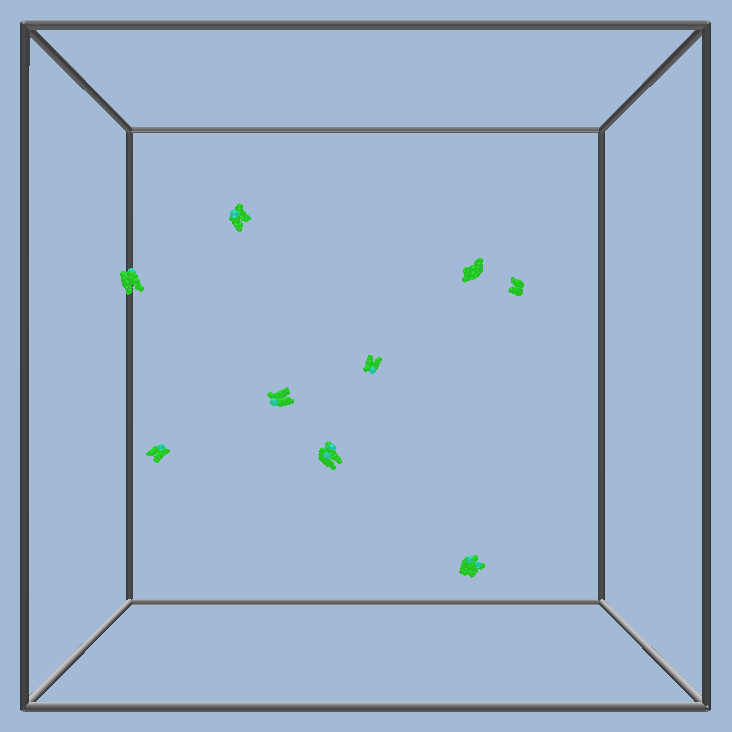
|
|
and the self assembled structures viewed.

|
|