|
| The simulator comes with several sample simulations. These are described below. |
|
| |
|
Post-synaptic Density
|
|
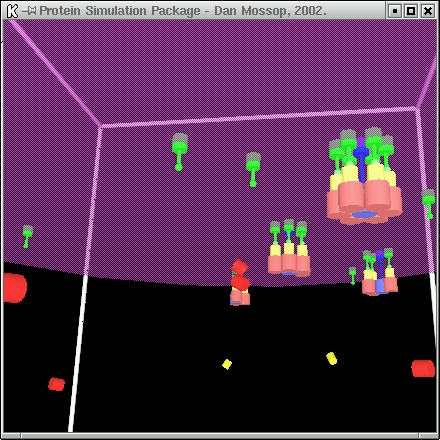
| This simulation models the formation of the post-synaptic density. The simulation proceeds as follows: |
|
-
The NMDA receptors (blue) are initially free from bonds, though bound to the membrane and can therefore only diffuse in two dimensions.
|
-
The CaMKII units (red) begin to bond to the base of the NMDA receptors.
|
-
This catalyses the bond between the AMPAR scaffolding proteins (yellow) and the CaMKII units.
|
-
The membrane-bounded AMPA units (green) then bond to the AMPAR proteins.
|
|
| When six AMPA units associate with an NMDA receptor, that channel considered fully formed and does not take part in any more reactions. |
|
|
| The post-synaptic density simulation is currently made available in three forms having one, four and eight channels respectively (synapse.obj, synapse4.obj
and synapse8.obj). Each can use either the synapse_clustered.tmpl or the synapse_unclustered.tmpl template files (the first forms clusters of channels, the
second keeps them separate). |
|
|
| |
|
Vesicle Coating
|
|
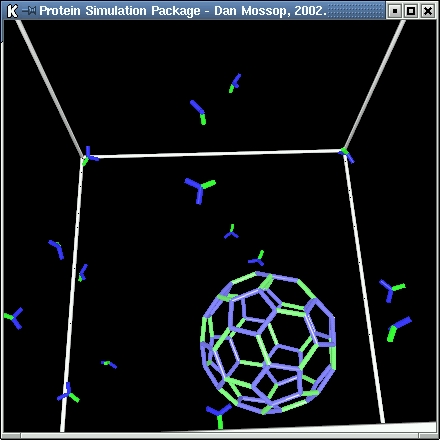
| In this simulation the formation of spherical vesicle coatings from individual clathrin proteins is modelled.
In the model the clathrin proteins have three 'legs' and are symmetrical along the first of these (green). The second and third legs are shown in blue.
The following rules govern the formation: |
|
-
If two clathrin proteins have no other bonds, there is a low probability of them forming a bond.
|
-
If either of two clathrin proteins have other bonds, there is a high probability of them forming a bond (unless no suitable bond-site pairing exists).
|
-
The bond-site on the end of the first leg on one clathrin protein can bond with the first leg of another.
|
-
The bond-site on the end of the second leg on one clathrin protein can bond with the third leg of another.
|
|
| When a vesicle becomes fully formed no more bonds can be made between the clathrin it contains and free-floating clathrin molecules.
The vesicles formed are of a fixed size with a regular geometric form in which each vertex of the structure marks the join between two
hexagons and a pentagon. |
|
|
| The vesicle simulation is available in two forms, one having enough for about one and a half vesicles (vesicle.obj) and the other having enough for about three
(vesicle2.obj). Both can use either the vesicle_hub.tmpl or the vesicle_no_hub.tmpl template files (the first contains a sphere at the centre of each clathrin and
is more polished visually, the second is faster).
Note that in some runs of the simulation many fragments of vesicles form, rather than the few complete structures. |
|
|
| |
|
Microtubule
|
|
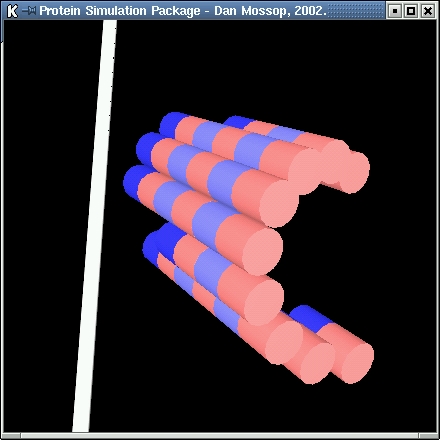
| In this simulation 13-protofilament microtubules are modelled. The microtubules are formed from alpha-beta tubulin subunits. These subunits can
join end-to-end and side-by-side. The lateral joins are staggered slightly resulting in a helical layout of subunits. |
|
|
| The microtubule simulation can be loaded by selecting microtubule.tmpl and microtubule.obj as the template files and object files respectively. |
|
|
|